Mitochondrial DNA Analysis
The National Geographic Society, IBM, geneticist Spencer Wells, and the Waitt Family Foundation are sponsoring a project to trace an individual's deep ancestry through DNA analysis of easily obtained cheek swab specimens. I ran this analysis on my mother Naomi's mitochondrial DNA (mtDNA), which would be the same as mine, as it is inherited solely through the maternal line. The results have been archived here from the National Geographic DNA analysis page. I have slightly modified the text and the map to improve readibility, and added some of my own explanatory text as well as supplementary material from Wikimedia commons sources.
Most people are familiar with the concept of DNA, the long molecules present in the center (the nucleus) of each and every one of our cells that contain the "instructions" for making a particular type of cell. Whether the cell is part of the liver, the brain, the muscle or a "stem cell" (with the potential to develop in a number of ways), it has the same set of instructions, only using those which are necessary for the task at hand. Every cell of an individual human - or plant or animal, for that matter - contains exactly the same DNA (with the exception of tumor cells, representing potentially lethal changes in the instruction sequence). The instructions are coded by specific arrangements of four types of subunits in the DNA molecule known as nucleides, which pair up on adjacent strands of DNA: adenine (A) pairing with guanine (G), and cytosine (C) pairing with thymine (T).
In addition to the nucleus of the cell (the part in the center with the DNA instructions), cells contain small structures known as mitochondria. These are involved in cellular energy production, but interestingly also contain DNA (mtDNA). One possible explanation for this is that these were long ago independant single celled organisms, which found an evolutionary advantage to live as "parasites" inside other larger organisms. However, these would not be true parasites, but rather examples of a symbiotic relationship, with the mitochondria and the host cell each providing an advantage to the other. When these larger cells evolved into multicellular organisms (such as you and me), the mitochondria came along as well. The mtDNA molecule is much smaller than the nuclear DNA.
The project traces markers (haplogroups) in mitochondrial DNA. Mitochondrial DNA (mtDNA) is useful for tracking maternal heritage, since it is passed on from mother to child virtually unchanged. If passed to a daughter, that person's children will also have the same mtDNA. There are occasional mutations, which are the sources of these various haplogroups that allow us to track migration patterns over thousands of years. The same thing can be done with the Y chromosome, which traces paternal heritage. However, only males can have paternal ancestry tracing done (since females do not have Y chromosomes). Any person can have an mtDNA analysis, since we all have mtDNA which we inherited from our mothers.
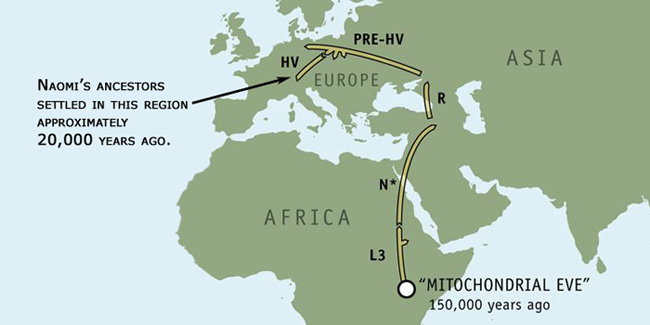
The first map is from the National Geographic project. It traces the migratory path taken by Naomi's maternal ancestors up to the point of divergence of the most recent subdivision, haplogroup HV, about 20,000 years ago.
The second map is from the Wikimedia commons, and shows the other known paths of human migration, all of which began with a woman who lived 150,000 years ago in Africa, and who was the most recent common matrilineal ancestor of all living humans.
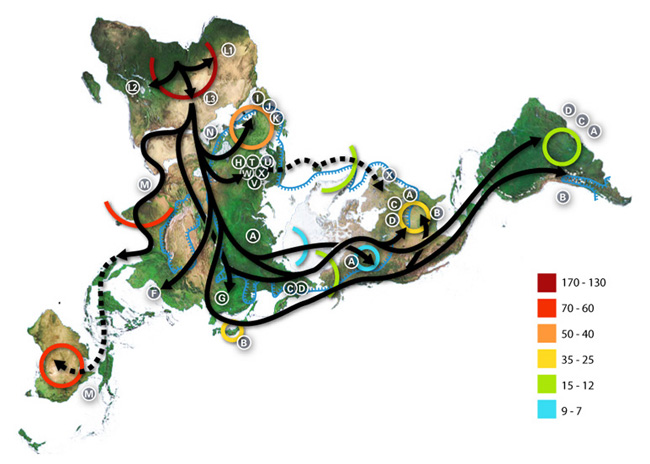
The numbers refer to thousand of years before the present. The letters are the various mtDNA haplogroups, whose geographic isolation is the basis for migratory tracing:
- Southern European: J, K
- Northern European: H, T, U, V, X
- Middle Eastern: J, N
- African: L, L1, L2, L3, L3*
- Asian: A, B, C, D, E, F, G (note: M is composed of C, D, E, and G)
- Native American: A, B, C, D, and sometimes X
Naomi's Genetic Sequence
Type: mtDNAHaplogroup: HV
Naomi's Mitochondrial HVR I Sequence
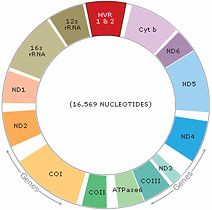
Above is displayed the sequence of Naomi's mitochondrial genome that was analyzed in the laboratory. Her sequence is compared against the Cambridge Reference Sequence (CRS), which is the standard mitochondrial sequence initially determined by researchers at Cambridge, UK. The differences between her DNA and the CRS are highlighted, and this mutation data allows researchers to reconstruct the migratory paths of her genetic lineage.
There are four types of these mutations:
- Substitution (transition): a nucleotide base mutation in which a pyrimidine base (C or T) is exchanged for another pyrimidine, or a purine base (A or G) is replaced with another purine. This is the most common type of single point mutation.
- Substitution (transversion): a base substitution in which a pyrimidine base (C or T) is exchanged for a purine base (A or G), or vice versa.
- Insertion: a mutation caused by the insertion of at least one extra nucleotide base in the DNA sequence.
- Deletion: a mutation caused by the deletion of at least one extra nucleotide base from the DNA sequence.
Naomi's Genetic History
Naomi's mtDNA results identify her as a member of haplogroup HV. This haplogroup is the final destination of a genetic journey that began some 150,000 years ago with an ancient mtDNA haplogroup called L3.
Haplogroup L3 occurs only in Africa, but on that continent its derivatives are found nearly everywhere. L3's subclades are most prevalent in East Africa.
This ancient lineage reflects an early divergence from humanity's common genetic coalescence point.
"Mitochondrial Eve," the common ancestor of all living humans, was born in Africa some 150,000 years ago. All existing MtDNA diversity began with Eve and it remains greatest, and subsequently oldest, in Africa.
Y chromosome polymorphisms on the male line of descent also point to an African origin for all humans, but our male common ancestor, "Adam," lived only about 60,000 years ago.
MtDNA and the Y chromosome are independent parts of our genetic makeup and each tells a different tale of successive genetic mutations over the eons. That is why their approximate coalescence points are different. Yet while the dates vary, both paths point emphatically to a surprisingly recent African origin for all humans.
The oldest known fossil remains of anatomically modern humans were found in Ethiopia's Omo River Valley. The skeletons, known as Omo I and Omo II, have been dated to about 195,000 years ago.
Although haplogroup L3 does not appear outside of Africa it is an important part of the human migrations from that continent to the rest of the world.
A single person of the L3 lineage gave rise to the M and N haplogroups some 80,000 years ago.
All Eurasian mtDNA lineages are subsequently descended from these two groups.
The African Ice Age was characterized by drought rather than by cold. But about 50,000 years ago a period of warmer temperatures and moist climate made even parts of the arid Sahara habitable. The climatic shift likely spurred hunter-gatherer migrations into a steppe-like Sahara-and beyond.
This "Saharan Gateway" led humans out of Africa to the Middle East. The route they took is uncertain. They may have traveled north down the Nile to the Mediterranean coast and the Sinai. Alternatively, they may have crossed what was then a land bridge connecting the Bab al Mandab to Arabia, after which they either skirted the then-lush, verdant eastern coast of the Red Sea or headed east along the Gulf of Aden towards the Arabian Sea.
When the climate again turned arid, expanding Saharan sands slammed the Saharan Gateway shut. The desert was at its driest between 20,000 and 40,000 years ago, and during this period Middle East migrants became isolated from Africa.
From their new Middle East location, however, they would go on to populate much of the world.
N is a macro-haplogroup descended from the African lineage L3. This line of descent, with haplogroup M, traces the first human migrations out of Africa. The ancient members of haplogroup N spawned sublineages found across Eurasia and, eventually, the Americas.
Early members of this group lived in the eastern Mediterranean and Near East region, where they likely coexisted for a time with pre-modern hominids such as Neandertals. Excavations in Israel's Kebara cave (Mount Carmel) have unearthed Neandertal skeletons at least as recent as 60,000 years old.
Growing cognitive abilities likely gave these Upper Paleolithic humans tremendous social advantages, evidenced by the appearance of modern thought and behavior. This "great leap forward" may have enabled our ancestors to outcompete and eventually replace evolutionary dead-end lineages such as Neandertals.
The macro-haplogroup N is composed of many subclades, which are often geographically distinct.
Haplogroup R is descended from N and has since dispersed across much of the globe. The lineage, in its many subgroups, appears on all continents except Australia and Antarctica.
Subgroups preHV, U, T, and J are found in Europe and the Near East. The R5 and R6 lineages arose on the Indian subcontinent.
Some 20,000 years ago, before the dawn of agriculture, the HV lineage descended from R and subsequently spread through Europe. Today, the haplogroup is more common in western Europe than in the east.
Learning more about such lineages will add further clarity to the big picture of human genetic diversity, and is a primary goal of the Genographic Project.